bpRNA-1m¶
bpRNA-1m is a database of single molecule secondary structures annotated using bpRNA.
Disclaimer¶
This is an UNOFFICIAL release of the bpRNA-1m by Center for Quantitative Life Sciences of the Oregon State University.
The team releasing bpRNA-1m did not write this dataset card for this dataset so this dataset card has been written by the MultiMolecule team.
Dataset Description¶
- Homepage: https://multimolecule.danling.org/datasets/bprna
- datasets: https://huggingface.co/datasets/multimolecule/bprna
- Point of Contact: Center for Quantitative Life Sciences of the Oregon State University
- Original URL: https://bprna.cgrb.oregonstate.edu/index.html
Example Entry¶
| id | sequence | secondary_structure | structural_annotation | functional_annotation |
|---|---|---|---|---|
| bpRNA_RFAM_1016 | AUUGCUUCUCGGCCUUUUGGCUAACAUCAAGU… | ......(((.((((....)))).)))...... | EEEEEESSSISSSSHHHHSSSSISSSXXXXXX… | NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN… |
Column Description¶
The converted dataset consists of the following columns, each providing specific information about the RNA secondary structures, consistent with the bpRNA standard:
-
id: A unique identifier for each RNA entry. This ID is derived from the original
.stafile name and serves as a reference to the specific RNA structure within the dataset. -
sequence: The nucleotide sequence of the RNA molecule, represented using the standard RNA bases:
- A: Adenine
- C: Cytosine
- G: Guanine
- U: Uracil
-
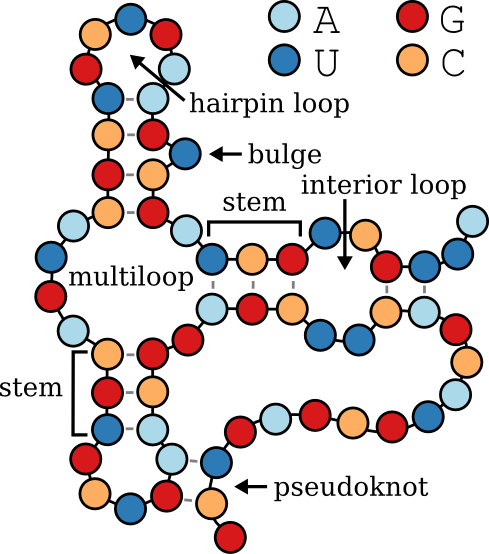
secondary_structure: The secondary structure of the RNA represented in dot-bracket notation, using up to three types of symbols to indicate base pairing and unpaired regions, as per bpRNA’s standard:
- Dots (
.): Represent unpaired nucleotides. - Parentheses (
(and)): Represent base pairs in standard stems (page 1). - Square Brackets (
[and]): Represent base pairs in pseudoknots (page 2). - Curly Braces (
{and}): Represent base pairs in additional pseudoknots (page 3).
- Dots (
-
structural_annotation: Structural annotations categorizing different regions of the RNA based on their roles within the secondary structure, consistent with bpRNA standards:
- E: External Loop – Regions that are unpaired and external to any loop or helix.
- S: Stem – Paired regions forming helical structures.
- H: Hairpin Loop – Unpaired regions at the end of a stem, forming a loop.
- I: Internal Loop – Unpaired regions between two stems.
- M: Multi-loop – Junctions where three or more stems converge.
- B: Bulge – Unpaired nucleotides on one side of a stem.
- X: Ambiguous or Undetermined – Regions where the structure is unknown or cannot be classified.
- K: Pseudoknot – Regions involved in pseudoknots, where base pairs cross each other.
-
functional_annotation: Functional annotations indicating specific functional elements or regions within the RNA sequence, as defined by bpRNA:
- N: None – No specific functional annotation is assigned.
- K: Pseudoknot – Marks nucleotides involved in pseudoknot structures, which can be functionally significant.
Variations¶
This dataset is available in two variants:
- bpRNA-1m: The main bpRNA-1m dataset.
- bpRNA-1m(90): bpRNA-1m(90) is a subset of bpRNA-1m containing RNAs with less than 90% sequence similarity.
Related Datasets¶
- bpRNA-spot: A subset of bpRNA-1m that applies CD-HIT (CD-HIT-EST) to remove sequences with more than 80% sequence similarity from bpRNA-1m.
- bpRNA-new: A dataset of newly discovered RNA families from Rfam 14.2, designed for cross-family validation to assess generalization capability.
License¶
This dataset is licensed under the AGPL-3.0 License.
| Text Only | |
|---|---|